Extracellular Vesicle Identification Using Label-Free Surface-Enhanced Raman Spectroscopy: Detection and Signal Analysis Strategies
Abstract
:1. Introduction
2. Characteristics of EVs and Their SERS Signals
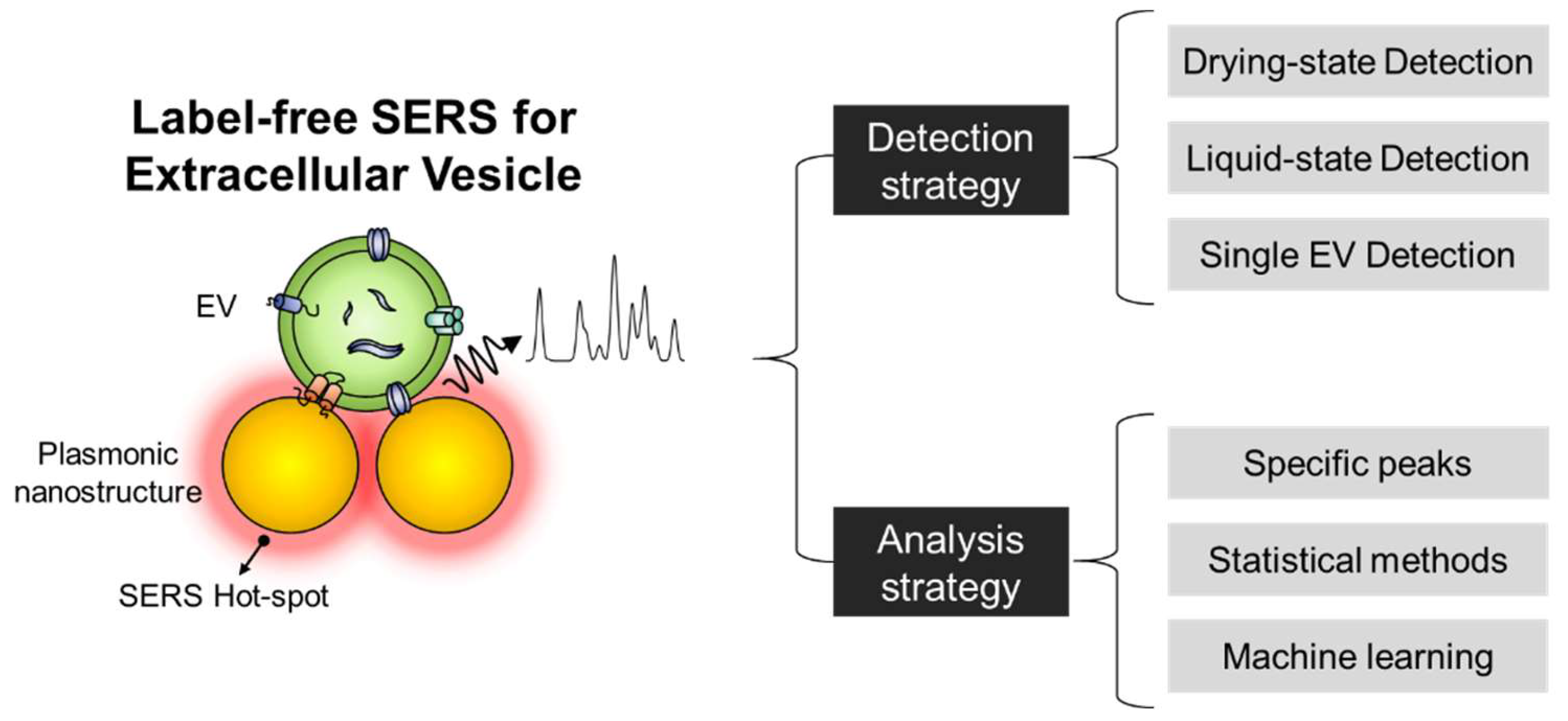
3. Label-Free SERS to Identify EVs
3.1. EV Detection Methods Using Label-Free SERS Substrate
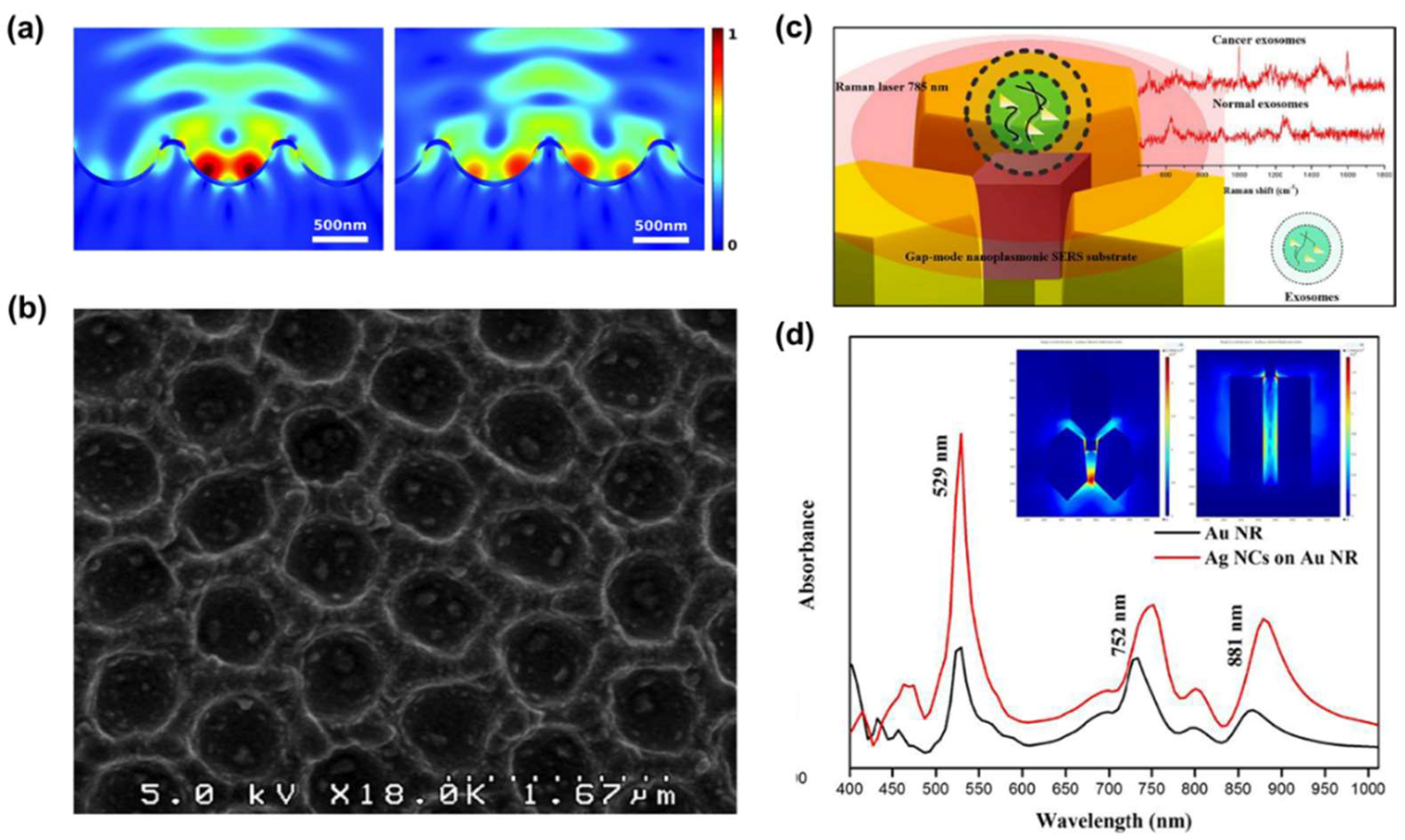
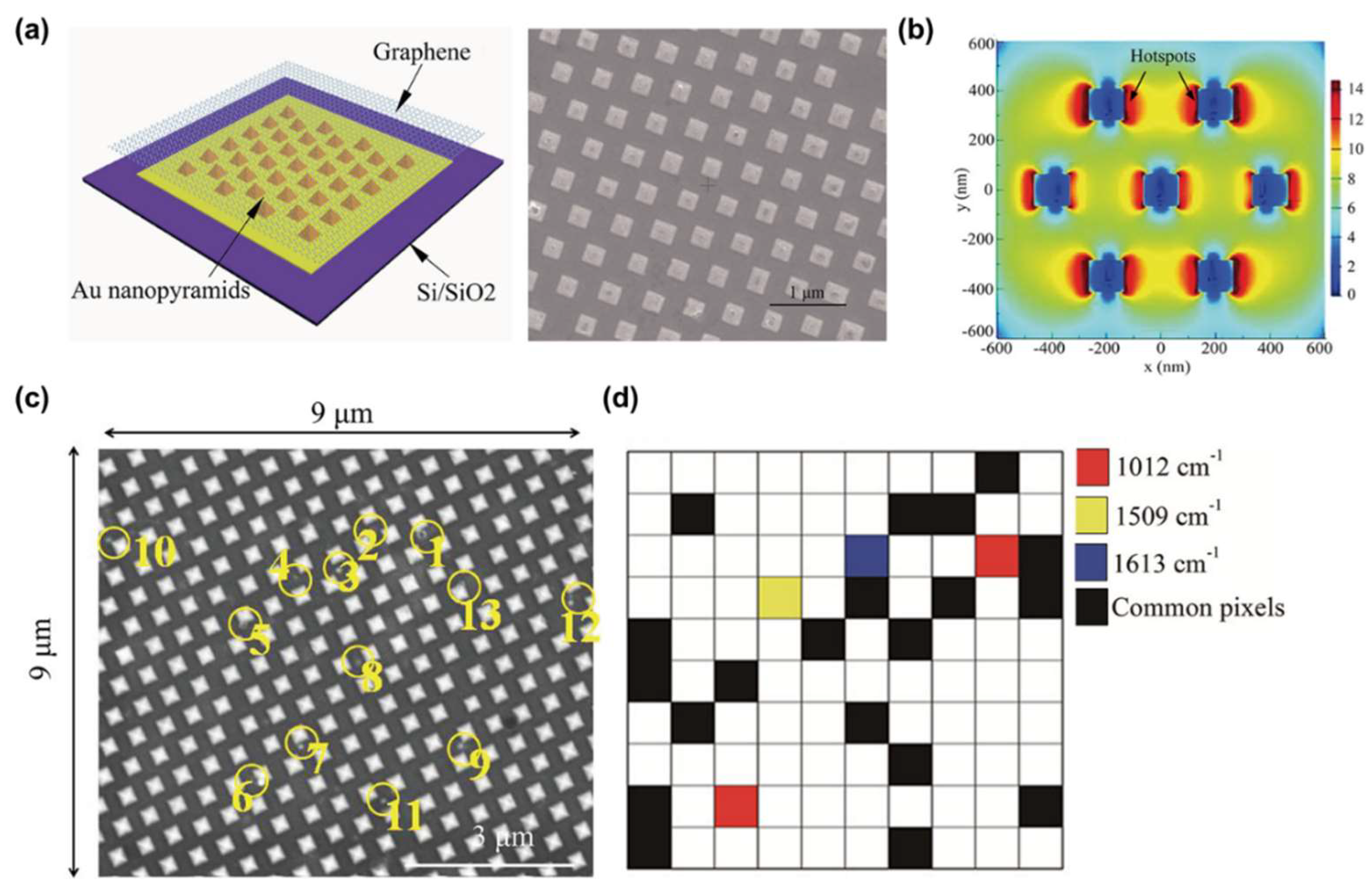
3.1.1. Drying-State Detection Methods
3.1.2. Liquid-State Detection Methods
3.2. Signal Analysis of the Label-Free SERS of EVs
3.2.1. Conventional Method
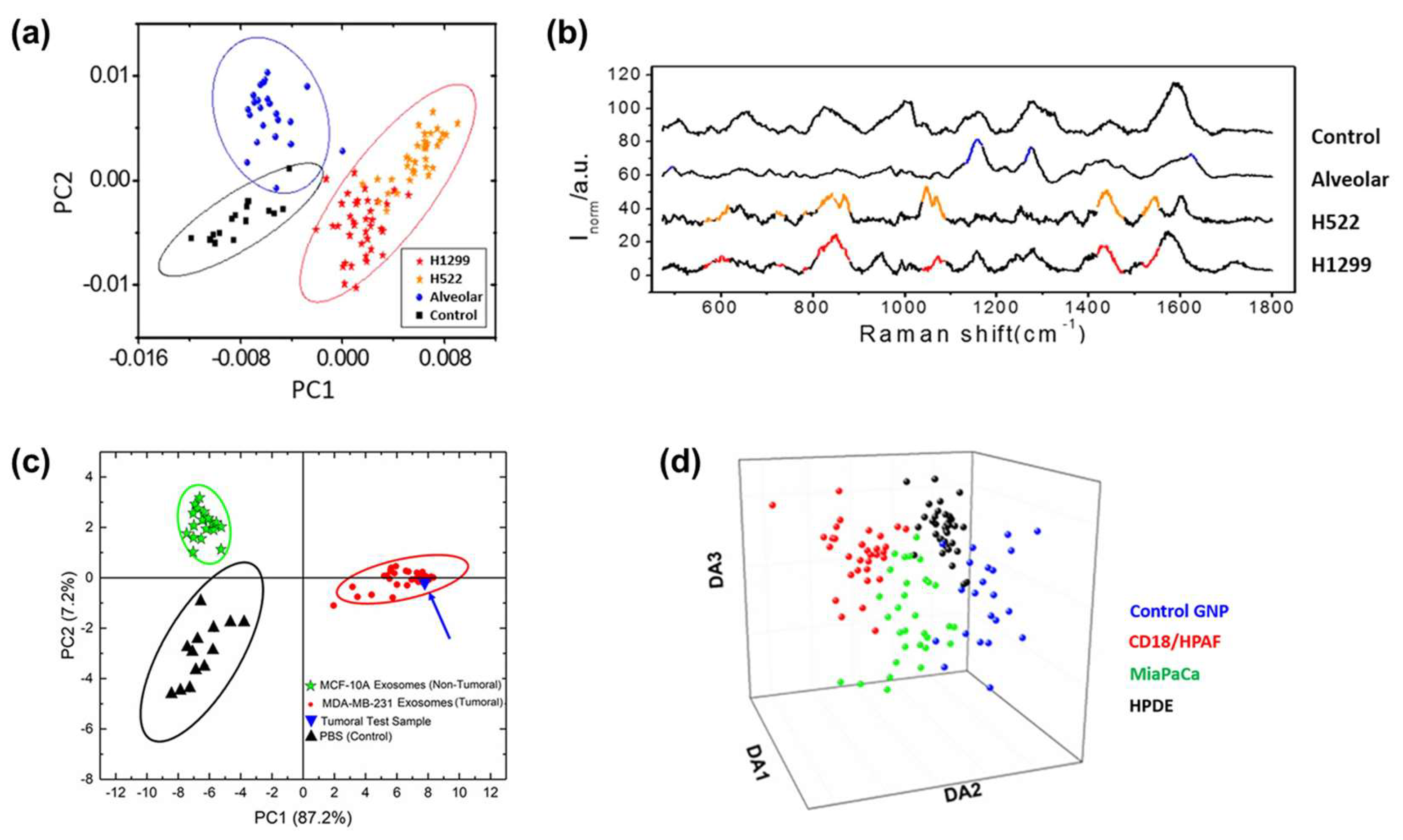
3.2.2. Principal Component Analysis (PCA)
3.2.3. Partial Least Square Discriminant Analysis (PLS-DA)
4. Challenges and Prospects of Label-Free SERS for EVs
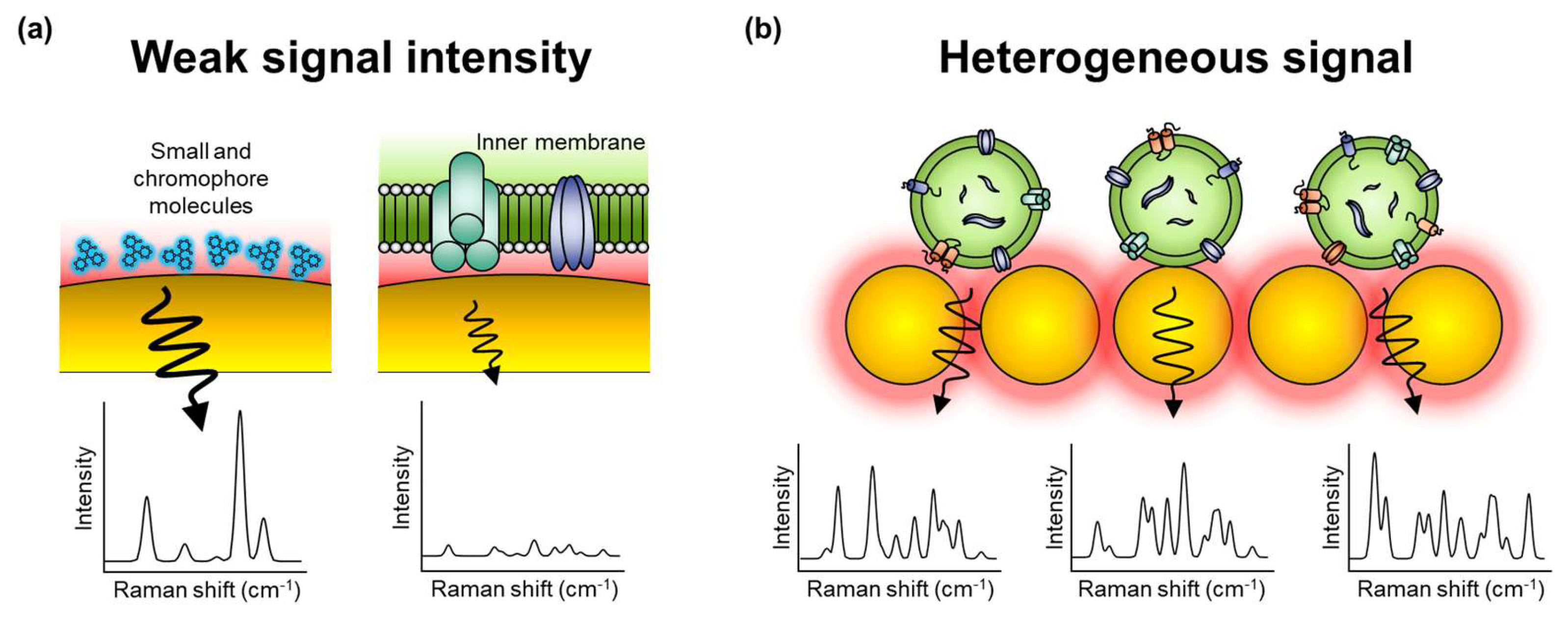
4.1. Challenges of Label-Free SERS for EVs
4.2. Prospects for Label-Free SERS for EVs
4.2.1. Prospect of the Detection Methods
4.2.2. Prospects of the Analysis Methods
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, eaau6977. [Google Scholar] [CrossRef] [PubMed]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De la Torre Gomez, C.; Goreham, R.V.; Bech Serra, J.J.; Nann, T.; Kussmann, M. “Exosomics”—A review of biophysics, biology and biochemistry of exosomes with a focus on human breast milk. Front. Genet. 2018, 9, 92. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Liu, Y.; Liu, H.; Tang, W.H. Exosomes: Biogenesis, biologic function and clinical potential. Cell Biosci. 2019, 9, 19. [Google Scholar] [CrossRef] [PubMed]
- Bell, B.M.; Kirk, I.D.; Hiltbrunner, S.; Gabrielsson, S.; Bultema, J.J. Designer exosomes as next-generation cancer immunotherapy. Nanomed. Nanotechnol. Biol. Med. 2016, 12, 163–169. [Google Scholar] [CrossRef]
- McBride, J.D.; Rodriguez-Menocal, L.; Badiavas, E.V. Extracellular vesicles as biomarkers and therapeutics in dermatology: A focus on exosomes. J. Investig. Dermatol. 2017, 137, 1622–1629. [Google Scholar] [CrossRef] [PubMed]
- Rojalin, T.; Phong, B.; Koster, H.J.; Carney, R.P. Nanoplasmonic approaches for sensitive detection and molecular characterization of extracellular vesicles. Front. Chem. 2019, 7, 279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rikkert, L.G.; Nieuwland, R.; Terstappen, L.; Coumans, F. Quality of extracellular vesicle images by transmission electron microscopy is operator and protocol dependent. J. Extracell. Vesicles 2019, 8, 1555419. [Google Scholar] [CrossRef] [PubMed]
- Soo, C.Y.; Song, Y.; Zheng, Y.; Campbell, E.C.; Riches, A.C.; Gunn-Moore, F.; Powis, S.J. Nanoparticle tracking analysis monitors microvesicle and exosome secretion from immune cells. Immunology 2012, 136, 192–197. [Google Scholar] [CrossRef] [Green Version]
- Vestad, B.; Llorente, A.; Neurauter, A.; Phuyal, S.; Kierulf, B.; Kierulf, P.; Skotland, T.; Sandvig, K.; Haug, K.B.F.; Øvstebø, R. Size and concentration analyses of extracellular vesicles by nanoparticle tracking analysis: A variation study. J. Extracell. Vesicles 2017, 6, 1344087. [Google Scholar] [CrossRef]
- Liao, G.; Liu, X.; Yang, X.; Wang, Q.; Geng, X.; Zou, L.; Liu, Y.; Li, S.; Zheng, Y.; Wang, K. Surface plasmon resonance assay for exosomes based on aptamer recognition and polydopamine-functionalized gold nanoparticles for signal amplification. Microchim. Acta 2020, 187, 251. [Google Scholar] [CrossRef]
- Sina, A.A.I.; Vaidyanathan, R.; Dey, S.; Carrascosa, L.G.; Shiddiky, M.J.; Trau, M. Real time and label free profiling of clinically relevant exosomes. Sci. Rep. 2016, 6, 30460. [Google Scholar] [CrossRef] [Green Version]
- Grasso, L.; Wyss, R.; Weidenauer, L.; Thampi, A.; Demurtas, D.; Prudent, M.; Lion, N.; Vogel, H. Molecular screening of cancer-derived exosomes by surface plasmon resonance spectroscopy. Anal. Bioanal. Chem. 2015, 407, 5425–5432. [Google Scholar] [CrossRef] [Green Version]
- Jones, R.R.; Hooper, D.C.; Zhang, L.; Wolverson, D.; Valev, V.K. Raman techniques: Fundamentals and frontiers. Nanoscale Res. Lett. 2019, 14, 1–34. [Google Scholar] [CrossRef] [Green Version]
- Shin, H.; Oh, S.; Kang, D.; Choi, Y. Protein Quantification and Imaging by Surface-Enhanced Raman Spectroscopy and Similarity Analysis. Adv. Sci. 2020, 7, 1903638. [Google Scholar] [CrossRef] [Green Version]
- Ding, S.-Y.; You, E.-M.; Tian, Z.-Q.; Moskovits, M. Electromagnetic theories of surface-enhanced Raman spectroscopy. Chem. Soc. Rev. 2017, 46, 4042–4076. [Google Scholar] [CrossRef] [PubMed]
- Choi, I.; Choi, Y. Plasmonic nanosensors: Review and prospect. IEEE J. Sel. Top. Quantum Electron. 2011, 18, 1110–1121. [Google Scholar] [CrossRef]
- Faust, O.; Hagiwara, Y.; Hong, T.J.; Lih, O.S.; Acharya, U.R. Deep learning for healthcare applications based on physiological signals: A review. Comput. Methods Programs Biomed. 2018, 161, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.; Wang, C.; Lu, L.; Wang, C.; Sun, Z.; Xiao, R. Dual-SERS biosensor for one-step detection of microRNAs in exosome and residual plasma of blood samples for diagnosing pancreatic cancer. Biosens. Bioelectron. 2019, 130, 204–213. [Google Scholar] [CrossRef]
- Zong, S.; Wang, L.; Chen, C.; Lu, J.; Zhu, D.; Zhang, Y.; Wang, Z.; Cui, Y. Facile detection of tumor-derived exosomes using magnetic nanobeads and SERS nanoprobes. Anal. Methods 2016, 8, 5001–5008. [Google Scholar] [CrossRef]
- Tian, Y.-F.; Ning, C.-F.; He, F.; Yin, B.-C.; Ye, B.-C. Highly sensitive detection of exosomes by SERS using gold nanostar@ Raman reporter@ nanoshell structures modified with a bivalent cholesterol-labeled DNA anchor. Analyst 2018, 143, 4915–4922. [Google Scholar] [CrossRef]
- Ning, C.-F.; Wang, L.; Tian, Y.-F.; Yin, B.-C.; Ye, B.-C. Multiple and sensitive SERS detection of cancer-related exosomes based on gold–silver bimetallic nanotrepangs. Analyst 2020, 145, 2795–2804. [Google Scholar] [CrossRef]
- Pang, Y.; Shi, J.; Yang, X.; Wang, C.; Sun, Z.; Xiao, R. Personalized detection of circling exosomal PD-L1 based on Fe3O4@ TiO2 isolation and SERS immunoassay. Biosens. Bioelectron. 2020, 148, 111800. [Google Scholar] [CrossRef]
- Lee, J.U.; Kim, W.H.; Lee, H.S.; Park, K.H.; Sim, S.J. Quantitative and Specific Detection of Exosomal miRNAs for Accurate Diagnosis of Breast Cancer Using a Surface-Enhanced Raman Scattering Sensor Based on Plasmonic Head-Flocked Gold Nanopillars. Small 2019, 15, 1804968. [Google Scholar] [CrossRef]
- Ma, D.; Huang, C.; Zheng, J.; Tang, J.; Li, J.; Yang, J.; Yang, R. Quantitative detection of exosomal microRNA extracted from human blood based on surface-enhanced Raman scattering. Biosens. Bioelectron. 2018, 101, 167–173. [Google Scholar] [CrossRef]
- Wang, Z.; Zong, S.; Wang, Y.; Li, N.; Li, L.; Lu, J.; Wang, Z.; Chen, B.; Cui, Y. Screening and multiple detection of cancer exosomes using an SERS-based method. Nanoscale 2018, 10, 9053–9062. [Google Scholar] [CrossRef]
- Kaufman, L.; Cooper, T.; Wallace, G.; Hawke, D.; Betts, D.; Hess, D.; Lagugné-Labarthet, F. Trapping and SERS identification of extracellular vesicles using nanohole arrays. In Plasmonics in Biology and Medicine XVI; International Society for Optics and Photonics: San Francisco, CA, USA, 2019; p. 108940B. [Google Scholar]
- Krafft, C.; Osei, E.B.; Popp, J.; Nazarenko, I. Raman and SERS spectroscopy for characterization of extracellular vesicles from control and prostate carcinoma patients. In Biomedical Vibrational Spectroscopy 2020: Advances in Research and Industry; International Society for Optics and Photonics: San Francisco, CA, USA, 2020; p. 1123602. [Google Scholar]
- Lee, C.; Carney, R.P.; Hazari, S.; Smith, Z.J.; Knudson, A.; Robertson, C.S.; Lam, K.S.; Wachsmann-Hogiu, S. 3D plasmonic nanobowl platform for the study of exosomes in solution. Nanoscale 2015, 7, 9290–9297. [Google Scholar] [CrossRef]
- Kwizera, E.A.; O’Connor, R.; Vinduska, V.; Williams, M.; Butch, E.R.; Snyder, S.E.; Chen, X.; Huang, X. Molecular detection and analysis of exosomes using surface-enhanced Raman scattering gold nanorods and a miniaturized device. Theranostics 2018, 8, 2722. [Google Scholar] [CrossRef]
- Lee, C.; Carney, R.; Lam, K.; Chan, J.W. SERS analysis of selectively captured exosomes using an integrin-specific peptide ligand. J. Raman Spectrosc. 2017, 48, 1771–1776. [Google Scholar] [CrossRef]
- Garcia-Rico, E.; Alvarez-Puebla, R.A.; Guerrini, L. Direct surface-enhanced Raman scattering (SERS) spectroscopy of nucleic acids: From fundamental studies to real-life applications. Chem. Soc. Rev. 2018, 47, 4909–4923. [Google Scholar] [CrossRef]
- Li, P.; Long, F.; Chen, W.; Chen, J.; Chu, P.K.; Wang, H. Fundamentals and applications of surface-enhanced Raman spectroscopy–based biosensors. Curr. Opin. Biomed. Eng. 2020, 13, 51–59. [Google Scholar] [CrossRef]
- Mercadal, P.A.; Encina, E.R.; Coronado, E.A. Colloidal SERS Substrate for the Ultrasensitive Detection of Biotinylated Antibodies Based on Near-Field Gradient within the Gap of Au Nanoparticle Dimers. J. Phys. Chem. C 2019, 123, 23577–23585. [Google Scholar] [CrossRef]
- Kamińska, A.; Winkler, K.; Kowalska, A.; Witkowska, E.; Szymborski, T.; Janeczek, A.; Waluk, J. Sers-based immunoassay in a microfluidic system for the multiplexed recognition of interleukins from blood plasma: Towards picogram detection. Sci. Rep. 2017, 7, 1–11. [Google Scholar] [CrossRef]
- Moon, J.; Yi, S.Y.; Hwang, A.; Eom, G.; Sim, J.; Jeong, J.; Lim, E.-K.; Chung, B.H.; Kim, B.; Jung, J. Facile and sensitive detection of influenza viruses using SERS antibody probes. RSC Adv. 2016, 6, 84415–84419. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.; Choi, N.; Jeon, J.H.; Rhie, G.-E.; Choo, J. SERS-based immunoassays for the detection of botulinum toxins A and B using magnetic beads. Sensors 2019, 19, 4081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pal, S.; Harmsen, S.; Oseledchyk, A.; Hsu, H.T.; Kircher, M.F. MUC1 aptamer targeted SERS nanoprobes. Adv. Funct. Mater. 2017, 27, 1606632. [Google Scholar] [CrossRef] [Green Version]
- Xu, X.; Ma, X.; Wang, H.; Wang, Z. Aptamer based SERS detection of Salmonella typhimurium using DNA-assembled gold nanodimers. Microchim. Acta 2018, 185, 325. [Google Scholar] [CrossRef]
- Díaz-Amaya, S.; Lin, L.-K.; Deering, A.J.; Stanciu, L.A. Aptamer-based SERS biosensor for whole cell analytical detection of E. coli O157: H7. Anal. Chim. Acta 2019, 1081, 146–156. [Google Scholar]
- Paulaitis, M.; Agarwal, K.; Nana-Sinkam, P. Dynamic scaling of exosome sizes. Langmuir 2018, 34, 9387–9393. [Google Scholar] [CrossRef] [Green Version]
- Lane, R.; Korbie, D.; Hill, M.; Trau, M. Extracellular vesicles as circulating cancer biomarkers: Opportunities and challenges. Clin. Transl. Med. 2018, 7, 14. [Google Scholar] [CrossRef]
- Welsh, J.A.; Holloway, J.A.; Wilkinson, J.S.; Englyst, N.A. Extracellular vesicle flow cytometry analysis and standardization. Front. Cell Dev. Biol. 2017, 5, 78. [Google Scholar] [CrossRef]
- Li, X.; Corbett, A.L.; Taatizadeh, E.; Tasnim, N.; Little, J.P.; Garnis, C.; Daugaard, M.; Guns, E.; Hoorfar, M.; Li, I.T. Challenges and opportunities in exosome research—Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019, 3, 011503. [Google Scholar] [CrossRef] [Green Version]
- Feliu, N.; Hassan, M.; Garcia Rico, E.; Cui, D.; Parak, W.; Alvarez-Puebla, R. SERS quantification and characterization of proteins and other biomolecules. Langmuir 2017, 33, 9711–9730. [Google Scholar] [CrossRef]
- Zheng, X.-S.; Jahn, I.J.; Weber, K.; Cialla-May, D.; Popp, J. Label-free SERS in biological and biomedical applications: Recent progress, current challenges and opportunities. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 197, 56–77. [Google Scholar] [CrossRef]
- Hamm, L.; Gee, A.; De Silva Indrasekara, A.S. Recent advancement in the surface-enhanced Raman spectroscopy-based biosensors for infectious disease diagnosis. Appl. Sci. 2019, 9, 1448. [Google Scholar] [CrossRef] [Green Version]
- Kruglik, S.G.; Royo, F.; Guigner, J.-M.; Palomo, L.; Seksek, O.; Turpin, P.-Y.; Tatischeff, I.; Falcón-Pérez, J.M. Raman tweezers microspectroscopy of circa 100 nm extracellular vesicles. Nanoscale 2019, 11, 1661–1679. [Google Scholar] [CrossRef] [Green Version]
- Sivashanmugan, K.; Huang, W.-L.; Lin, C.-H.; Liao, J.-D.; Lin, C.-C.; Su, W.-C.; Wen, T.-C. Bimetallic nanoplasmonic gap-mode SERS substrate for lung normal and cancer-derived exosomes detection. J. Taiwan Inst. Chem. Eng. 2017, 80, 149–155. [Google Scholar] [CrossRef]
- Dong, S.; Wang, Y.; Liu, Z.; Zhang, W.; Yi, K.; Zhang, X.; Zhang, X.; Jiang, C.; Yang, S.; Wang, F. Beehive-inspired macroporous SERS probe for cancer detection through capturing and analyzing exosomes in plasma. ACS Appl. Mater. Interfaces 2020, 12, 5136–5146. [Google Scholar] [CrossRef]
- Yan, Z.; Dutta, S.; Liu, Z.; Yu, X.; Mesgarzadeh, N.; Ji, F.; Bitan, G.; Xie, Y.-H. A label-free platform for identification of exosomes from different sources. ACS Sens. 2019, 4, 488–497. [Google Scholar] [CrossRef]
- Ferreira, N.; Marques, A.; Águas, H.; Bandarenka, H.; Martins, R.; Bodo, C.; Costa-Silva, B.; Fortunato, E. Label-free nanosensing platform for breast cancer exosome profiling. ACS Sens. 2019, 4, 2073–2083. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, L.; Fang, Y.; Zheng, D.; Lin, T.; Wang, H. Label-Free Exosomal Detection and Classification in Rapid Discriminating Different Cancer Types Based on Specific Raman Phenotypes and Multivariate Statistical Analysis. Molecules 2019, 24, 2947. [Google Scholar] [CrossRef] [Green Version]
- Carmicheal, J.; Hayashi, C.; Huang, X.; Liu, L.; Lu, Y.; Krasnoslobodtsev, A.; Lushnikov, A.; Kshirsagar, P.G.; Patel, A.; Jain, M. Label-free characterization of exosome via surface enhanced Raman spectroscopy for the early detection of pancreatic cancer. Nanomed. Nanotechnol. Biol. Med. 2019, 16, 88–96. [Google Scholar] [CrossRef] [PubMed]
- Shin, H.; Jeong, H.; Park, J.; Hong, S.; Choi, Y. Correlation between cancerous exosomes and protein markers based on surface-enhanced Raman spectroscopy (SERS) and principal component analysis (PCA). ACS Sens. 2018, 3, 2637–2643. [Google Scholar] [CrossRef] [PubMed]
- Stremersch, S.; Marro, M.; Pinchasik, B.E.; Baatsen, P.; Hendrix, A.; De Smedt, S.C.; Loza-Alvarez, P.; Skirtach, A.G.; Raemdonck, K.; Braeckmans, K. Identification of Individual Exosome-Like Vesicles by Surface Enhanced Raman Spectroscopy. Small 2016, 12, 3292–3301. [Google Scholar] [CrossRef]
- Fraire, J.C.; Stremersch, S.; Bouckaert, D.; Monteyne, T.; De Beer, T.; Wuytens, P.; De Rycke, R.; Skirtach, A.G.; Raemdonck, K.; De Smedt, S. Improved label-free identification of individual exosome-like vesicles with Au@ Ag nanoparticles as SERS substrate. ACS Appl. Mater. Interfaces 2019, 11, 39424–39435. [Google Scholar] [CrossRef]
- Shin, H.; Oh, S.; Hong, S.; Kang, M.; Kang, D.; Ji, Y.-g.; Choi, B.H.; Kang, K.-W.; Jeong, H.; Park, Y. Early-Stage Lung Cancer Diagnosis by Deep Learning-Based Spectroscopic Analysis of Circulating Exosomes. ACS Nano 2020, 14, 5435–5444. [Google Scholar] [CrossRef]
- Park, J.; Hwang, M.; Choi, B.; Jeong, H.; Jung, J.-H.; Kim, H.K.; Hong, S.; Park, J.-H.; Choi, Y. Exosome classification by pattern analysis of surface-enhanced Raman spectroscopy data for lung cancer diagnosis. Anal. Chem. 2017, 89, 6695–6701. [Google Scholar] [CrossRef] [PubMed]
- Chisanga, M.; Muhamadali, H.; Ellis, D.I.; Goodacre, R. Enhancing disease diagnosis: Biomedical applications of surface-enhanced Raman scattering. Appl. Sci. 2019, 9, 1163. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.-J.; Lei, Z.-C.; Li, J.; Zong, C.; Yang, C.J.; Ren, B. Label-free surface-enhanced Raman spectroscopy detection of DNA with single-base sensitivity. J. Am. Chem. Soc. 2015, 137, 5149–5154. [Google Scholar] [CrossRef]
- Harding, C.V.; Heuser, J.E.; Stahl, P.D. Exosomes: Looking back three decades and into the future. J. Cell Biol. 2013, 200, 367–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barros, F.M.; Carneiro, F.; Machado, J.C.; Melo, S.A. Exosomes and immune response in cancer: Friends or foes? Front. Immunol. 2018, 9, 730. [Google Scholar] [CrossRef] [PubMed]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [Green Version]
- Raimondo, S.; Saieva, L.; Corrado, C.; Fontana, S.; Flugy, A.; Rizzo, A.; De Leo, G.; Alessandro, R. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun. Signal. 2015, 13, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Zhang, T.; Zheng, M.; Liu, Y.; Chen, Z. Exosomal proteins as potential markers of tumor diagnosis. J. Hematol. Oncol. 2017, 10, 175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chun-Yan, L.; Zi-Yi, Z.; Tian-Lin, Y.; Yi-Li, W.; Bao, L.; Jiao, L.; Wei-Jun, D. Liquid biopsy biomarkers of renal interstitial fibrosis based on urinary exosome. Exp. Mol. Pathol. 2018, 105, 223–228. [Google Scholar] [CrossRef]
- Cheng, J.; Nonaka, T.; Wong, D.T. Salivary exosomes as nanocarriers for cancer biomarker delivery. Materials 2019, 12, 654. [Google Scholar] [CrossRef] [Green Version]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Shimoda, A.; Sawada, S.-i.; Sasaki, Y.; Akiyoshi, K. Exosome surface glycans reflect osteogenic differentiation of mesenchymal stem cells: Profiling by an evanescent field fluorescence-assisted lectin array system. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kleijmeer, M.J.; Stoorvogel, W.; Griffith, J.M.; Yoshie, O.; Geuze, H.J. Selective enrichment of tetraspan proteins on the internal vesicles of multivesicular endosomes and on exosomes secreted by human B-lymphocytes. J. Biol. Chem. 1998, 273, 20121–20127. [Google Scholar]
- Andreu, Z.; Yáñez-Mó, M. Tetraspanins in extracellular vesicle formation and function. Front. Immunol. 2014, 5, 442. [Google Scholar] [CrossRef] [Green Version]
- Wang, N.; Song, X.; Liu, L.; Niu, L.; Wang, X.; Song, X.; Xie, L. Circulating exosomes contain protein biomarkers of metastatic non-small-cell lung cancer. Cancer Sci. 2018, 109, 1701–1709. [Google Scholar] [CrossRef] [Green Version]
- Sandfeld-Paulsen, B.; Jakobsen, K.R.; Bæk, R.; Folkersen, B.H.; Rasmussen, T.R.; Meldgaard, P.; Varming, K.; Jørgensen, M.M.; Sorensen, B.S. Exosomal proteins as diagnostic biomarkers in lung cancer. J. Thorac. Oncol. 2016, 11, 1701–1710. [Google Scholar] [CrossRef] [Green Version]
- Liang, K.; Liu, F.; Fan, J.; Sun, D.; Liu, C.; Lyon, C.J.; Bernard, D.W.; Li, Y.; Yokoi, K.; Katz, M.H. Nanoplasmonic quantification of tumour-derived extracellular vesicles in plasma microsamples for diagnosis and treatment monitoring. Nat. Biomed. Eng. 2017, 1, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sandfeld-Paulsen, B.; Aggerholm-Pedersen, N.; Baek, R.; Jakobsen, K.; Meldgaard, P.; Folkersen, B.; Rasmussen, T.; Varming, K.; Jørgensen, M.; Sorensen, B. Exosomal proteins as prognostic biomarkers in non-small cell lung cancer. Mol. Oncol. 2016, 10, 1595–1602. [Google Scholar] [CrossRef] [Green Version]
- Kahlert, C.; Melo, S.A.; Protopopov, A.; Tang, J.; Seth, S.; Koch, M.; Zhang, J.; Weitz, J.; Chin, L.; Futreal, A. Identification of double-stranded genomic DNA spanning all chromosomes with mutated KRAS and p53 DNA in the serum exosomes of patients with pancreatic cancer. J. Biol. Chem. 2014, 289, 3869–3875. [Google Scholar] [CrossRef] [Green Version]
- Al-Nedawi, K.; Meehan, B.; Kerbel, R.S.; Allison, A.C.; Rak, J. Endothelial expression of autocrine VEGF upon the uptake of tumor-derived microvesicles containing oncogenic EGFR. Proc. Natl. Acad. Sci. USA 2009, 106, 3794–3799. [Google Scholar] [CrossRef] [Green Version]
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2017, 70, 122–132. [Google Scholar] [CrossRef]
- Kalluri, R.; LeBleu, V.S. Discovery of double-stranded genomic DNA in circulating exosomes. In Cold Spring Harbor Symposia on Quantitative Biology; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2016; pp. 275–280. [Google Scholar]
- Thakur, B.K.; Zhang, H.; Becker, A.; Matei, I.; Huang, Y.; Costa-Silva, B.; Zheng, Y.; Hoshino, A.; Brazier, H.; Xiang, J. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. 2014, 24, 766–769. [Google Scholar] [CrossRef] [Green Version]
- Yokoi, A.; Villar-Prados, A.; Oliphint, P.A.; Zhang, J.; Song, X.; De Hoff, P.; Morey, R.; Liu, J.; Roszik, J.; Clise-Dwyer, K. Mechanisms of nuclear content loading to exosomes. Sci. Adv. 2019, 5, eaax8849. [Google Scholar] [CrossRef] [Green Version]
- Dieringer, J.A.; McFarland, A.D.; Shah, N.C.; Stuart, D.A.; Whitney, A.V.; Yonzon, C.R.; Young, M.A.; Zhang, X.; Van Duyne, R.P. Introductory lecture surface enhanced Raman spectroscopy: New materials, concepts, characterization tools, and applications. Faraday Discuss. 2006, 132, 9–26. [Google Scholar] [CrossRef]
- Xu, L.-J.; Zong, C.; Zheng, X.-S.; Hu, P.; Feng, J.-M.; Ren, B. Label-free detection of native proteins by surface-enhanced Raman spectroscopy using iodide-modified nanoparticles. Anal. Chem. 2014, 86, 2238–2245. [Google Scholar] [CrossRef]
- Shao, F.; Lu, Z.; Liu, C.; Han, H.; Chen, K.; Li, W.; He, Q.; Peng, H.; Chen, J. Hierarchical nanogaps within bioscaffold arrays as a high-performance SERS substrate for animal virus biosensing. ACS Appl. Mater. Interfaces 2014, 6, 6281–6289. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, X.; Luo, C.; Liu, Z.; Chen, Y.; Dong, S.; Jiang, C.; Yang, S.; Wang, F.; Xiao, X. Volume-Enhanced Raman Scattering Detection of Viruses. Small 2019, 15, 1805516. [Google Scholar] [CrossRef] [PubMed]
- Beit-Yannai, E.; Tabak, S.; Stamer, W.D. Physical exosome: Exosome interactions. J. Cell. Mol. Med. 2018, 22, 2001–2006. [Google Scholar] [CrossRef] [Green Version]
- Soares Martins, T.; Catita, J.; Martins Rosa, I.; AB da Cruz e Silva, O.; Henriques, A.G. Exosome isolation from distinct biofluids using precipitation and column-based approaches. PLoS ONE 2018, 13, e0198820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, M.; Ye, J. Reproducibility in surface-enhanced Raman spectroscopy. J. Shanghai Jiaotong Univ. (Sci.) 2014, 19, 681–690. [Google Scholar] [CrossRef]
- Langer, J.; Jimenez de Aberasturi, D.; Aizpurua, J.; Alvarez-Puebla, R.A.; Auguié, B.; Baumberg, J.J.; Bazan, G.C.; Bell, S.E.; Boisen, A.; Brolo, A.G. Present and future of surface-enhanced Raman scattering. ACS Nano 2019, 14, 28–117. [Google Scholar] [CrossRef] [Green Version]
- Zong, C.; Xu, M.; Xu, L.-J.; Wei, T.; Ma, X.; Zheng, X.-S.; Hu, R.; Ren, B. Surface-enhanced Raman spectroscopy for bioanalysis: Reliability and challenges. Chem. Rev. 2018, 118, 4946–4980. [Google Scholar] [CrossRef]
- Theodoraki, M.N.; Hoffmann, T.K.; Whiteside, T.L. Separation of plasma-derived exosomes into CD3 (+) and CD3 (–) fractions allows for association of immune cell and tumour cell markers with disease activity in HNSCC patients. Clin. Exp. Immunol. 2018, 192, 271–283. [Google Scholar] [CrossRef] [Green Version]
- Lazzari, E.; Schena, T.; Marcelo, M.C.A.; Primaz, C.T.; Silva, A.N.; Ferrão, M.F.; Bjerk, T.; Caramão, E.B. Classification of biomass through their pyrolytic bio-oil composition using FTIR and PCA analysis. Ind. Crop. Prod. 2018, 111, 856–864. [Google Scholar] [CrossRef]
- Shinzawa, H.; Awa, K.; Kanematsu, W.; Ozaki, Y. Multivariate data analysis for Raman spectroscopic imaging. J. Raman Spectrosc. 2009, 40, 1720–1725. [Google Scholar] [CrossRef]
- Wang, X.; Paliwal, K.K. Feature extraction and dimensionality reduction algorithms and their applications in vowel recognition. Pattern Recognit. 2003, 36, 2429–2439. [Google Scholar] [CrossRef]
- Li, J.; Zhao, B.; Zhang, H. Face recognition based on PCA and LDA combination feature extraction. In Proceedings of the 2009 First International Conference on Information Science and Engineering, Nanjing, China, 26–28 December 2009; pp. 1240–1243. [Google Scholar]
- Chen, L.; Wang, Y.; Liu, N.; Lin, D.; Weng, C.; Zhang, J.; Zhu, L.; Chen, W.; Chen, R.; Feng, S. Near-infrared confocal micro-Raman spectroscopy combined with PCA–LDA multivariate analysis for detection of esophageal cancer. Laser Phys. 2013, 23, 065601. [Google Scholar] [CrossRef]
- Yang, J.; Yang, J.-Y. Why can LDA be performed in PCA transformed space? Pattern Recognit. 2003, 36, 563–566. [Google Scholar] [CrossRef]
- Hamasha, K.; Mohaidat, Q.I.; Putnam, R.A.; Woodman, R.C.; Palchaudhuri, S.; Rehse, S.J. Sensitive and specific discrimination of pathogenic and nonpathogenic Escherichia coli using Raman spectroscopy—A comparison of two multivariate analysis techniques. Biomed. Opt. Express 2013, 4, 481–489. [Google Scholar] [CrossRef] [Green Version]
- Lee, L.C.; Liong, C.-Y.; Jemain, A.A. Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: A review of contemporary practice strategies and knowledge gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef]
- Nguyen, D.V.; Rocke, D.M. Tumor classification by partial least squares using microarray gene expression data. Bioinformatics 2002, 18, 39–50. [Google Scholar] [CrossRef] [Green Version]
- Nocairi, H.; Qannari, E.M.; Vigneau, E.; Bertrand, D. Discrimination on latent components with respect to patterns. Application to multicollinear data. Comput. Stat. Data Anal. 2005, 48, 139–147. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. J. Chemom. Soc. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Smolarz, M.; Pietrowska, M.; Matysiak, N.; Mielańczyk, Ł.; Widłak, P. Proteome profiling of exosomes purified from a Small amount of human serum: The problem of Co-purified serum components. Proteomes 2019, 7, 18. [Google Scholar] [CrossRef] [Green Version]
- Zitvogel, L.; Fernandez, N.; Lozier, A.; Wolfers, J.; Regnault, A.; Raposo, G.; Amigorena, S. Dendritic cells or their exosomes are effective biotherapies of cancer. Eur. J. Cancer 1999, 35, S36–S38. [Google Scholar] [CrossRef]
- Tao, S.-C.; Guo, S.-C.; Zhang, C.-Q. Platelet-derived extracellular vesicles: An emerging therapeutic approach. Int. J. Biol. Sci. 2017, 13, 828. [Google Scholar] [CrossRef] [Green Version]
- Barik, A.; Cherukulappurath, S.; Wittenberg, N.J.; Johnson, T.W.; Oh, S.-H. Dielectrophoresis-assisted Raman spectroscopy of intravesicular analytes on metallic pyramids. Anal. Chem. 2016, 88, 1704–1710. [Google Scholar] [CrossRef]
- Wu, M.; Ouyang, Y.; Wang, Z.; Zhang, R.; Huang, P.-H.; Chen, C.; Li, H.; Li, P.; Quinn, D.; Dao, M. Isolation of exosomes from whole blood by integrating acoustics and microfluidics. Proc. Natl. Acad. Sci. USA 2017, 114, 10584–10589. [Google Scholar] [CrossRef] [Green Version]
- Deng, C.; Naler, L.B.; Lu, C. Microfluidic epigenomic mapping technologies for precision medicine. Lab Chip 2019, 19, 2630–2650. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Fraser, K.; Ghaddar, B.; Yang, K.; Kim, E.; Balaj, L.; Chiocca, E.A.; Breakefield, X.O.; Lee, H.; Weissleder, R. Multiplexed profiling of single extracellular vesicles. ACS Nano 2018, 12, 494–503. [Google Scholar] [CrossRef]
- Liu, C.; Xu, X.; Li, B.; Situ, B.; Pan, W.; Hu, Y.; An, T.; Yao, S.; Zheng, L. Single-exosome-counting immunoassays for cancer diagnostics. Nano Lett. 2018, 18, 4226–4232. [Google Scholar] [CrossRef]
- Han, B.; Zhang, Y.-L.; Zhu, L.; Chen, X.-H.; Ma, Z.-C.; Zhang, X.-L.; Wang, J.-N.; Wang, W.; Liu, Y.-Q.; Chen, Q.-D. Direct laser scribing of AgNPs@ RGO biochip as a reusable SERS sensor for DNA detection. Sens. Actuators B Chem. 2018, 270, 500–507. [Google Scholar] [CrossRef]
- Villa, J.E.; Poppi, R.J. A portable SERS method for the determination of uric acid using a paper-based substrate and multivariate curve resolution. Analyst 2016, 141, 1966–1972. [Google Scholar] [CrossRef] [Green Version]
- Carney, R.P.; Hazari, S.; Colquhoun, M.; Tran, D.; Hwang, B.; Mulligan, M.S.; Bryers, J.D.; Girda, E.; Leiserowitz, G.S.; Smith, Z.J. Multispectral optical tweezers for biochemical fingerprinting of CD9-positive exosome subpopulations. Anal. Chem. 2017, 89, 5357–5363. [Google Scholar] [CrossRef] [Green Version]
- Smith, Z.J.; Lee, C.; Rojalin, T.; Carney, R.P.; Hazari, S.; Knudson, A.; Lam, K.; Saari, H.; Ibañez, E.L.; Viitala, T. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J. Extracell. Vesicles 2015, 4, 28533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tatischeff, I.; Larquet, E.; Falcón-Pérez, J.M.; Turpin, P.-Y.; Kruglik, S.G. Fast characterisation of cell-derived extracellular vesicles by nanoparticles tracking analysis, cryo-electron microscopy, and Raman tweezers microspectroscopy. J. Extracell. Vesicles 2012, 1, 19179. [Google Scholar] [CrossRef]
- Dai, Y.; Bai, S.; Hu, C.; Chu, K.; Shen, B.; Smith, Z.J. Combined morpho-chemical profiling of individual extracellular vesicles and functional nanoparticles without labels. Anal. Chem. 2020, 92, 5585–5594. [Google Scholar] [CrossRef]
- Lee, W.; Lenferink, A.T.; Otto, C.; Offerhaus, H.L. Classifying Raman spectra of extracellular vesicles based on convolutional neural networks for prostate cancer detection. J. Raman Spectrosc. 2020, 51, 293–300. [Google Scholar] [CrossRef]
- Li, X.; Liu, L.; Zhou, J.; Wang, C. Heterogeneity analysis and diagnosis of complex diseases based on deep learning method. Sci. Rep. 2018, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.L.; Mahjoubfar, A.; Tai, L.-C.; Blaby, I.K.; Huang, A.; Niazi, K.R.; Jalali, B. Deep learning in label-free cell classification. Sci. Rep. 2016, 6, 21471. [Google Scholar] [CrossRef] [Green Version]
- Lussier, F.; Thibault, V.; Charron, B.; Wallace, G.Q.; Masson, J.-F. Deep learning and artificial intelligence methods for Raman and surface-enhanced Raman scattering. Trends Anal. Chem. 2020, 124, 115796. [Google Scholar] [CrossRef]
- Yan, H.; Yu, M.; Xia, J.; Zhu, L.; Zhang, T.; Zhu, Z. Tongue squamous cell carcinoma discrimination with Raman spectroscopy and convolutional neural networks. Vib. Spectrosc. 2019, 103, 102938. [Google Scholar] [CrossRef]
- Li, X.; Yang, T.; Li, S.; Jin, L.; Wang, D.; Guan, D.; Ding, J. Noninvasive liver diseases detection based on serum surface enhanced Raman spectroscopy and statistical analysis. Opt. Express 2015, 23, 18361–18372. [Google Scholar] [CrossRef]
- Guevara, E.; Torres-Galván, J.C.; Ramírez-Elías, M.G.; Luevano-Contreras, C.; González, F.J. Use of Raman spectroscopy to screen diabetes mellitus with machine learning tools. Biomed. Opt. Express 2018, 9, 4998–5010. [Google Scholar] [CrossRef]
- Thrift, W.J.; Cabuslay, A.; Laird, A.B.; Ranjbar, S.; Hochbaum, A.I.; Ragan, R. Surface-enhanced Raman scattering-based odor compass: Locating multiple chemical sources and pathogens. ACS Sens. 2019, 4, 2311–2319. [Google Scholar] [CrossRef]
- Khan, S.; Ullah, R.; Khan, A.; Wahab, N.; Bilal, M.; Ahmed, M. Analysis of dengue infection based on Raman spectroscopy and support vector machine (SVM). Biomed. Opt. Express 2016, 7, 2249–2256. [Google Scholar] [CrossRef]
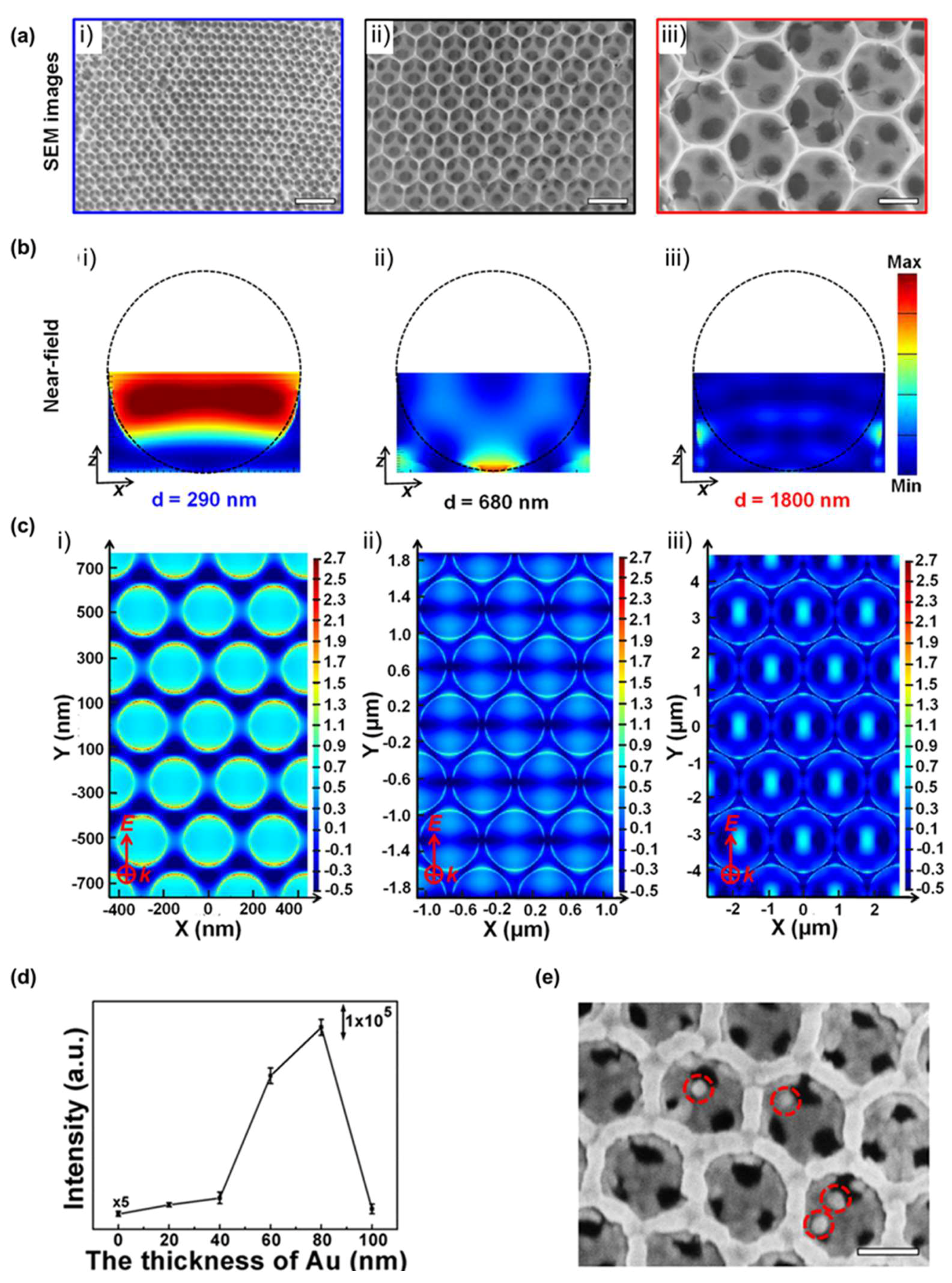
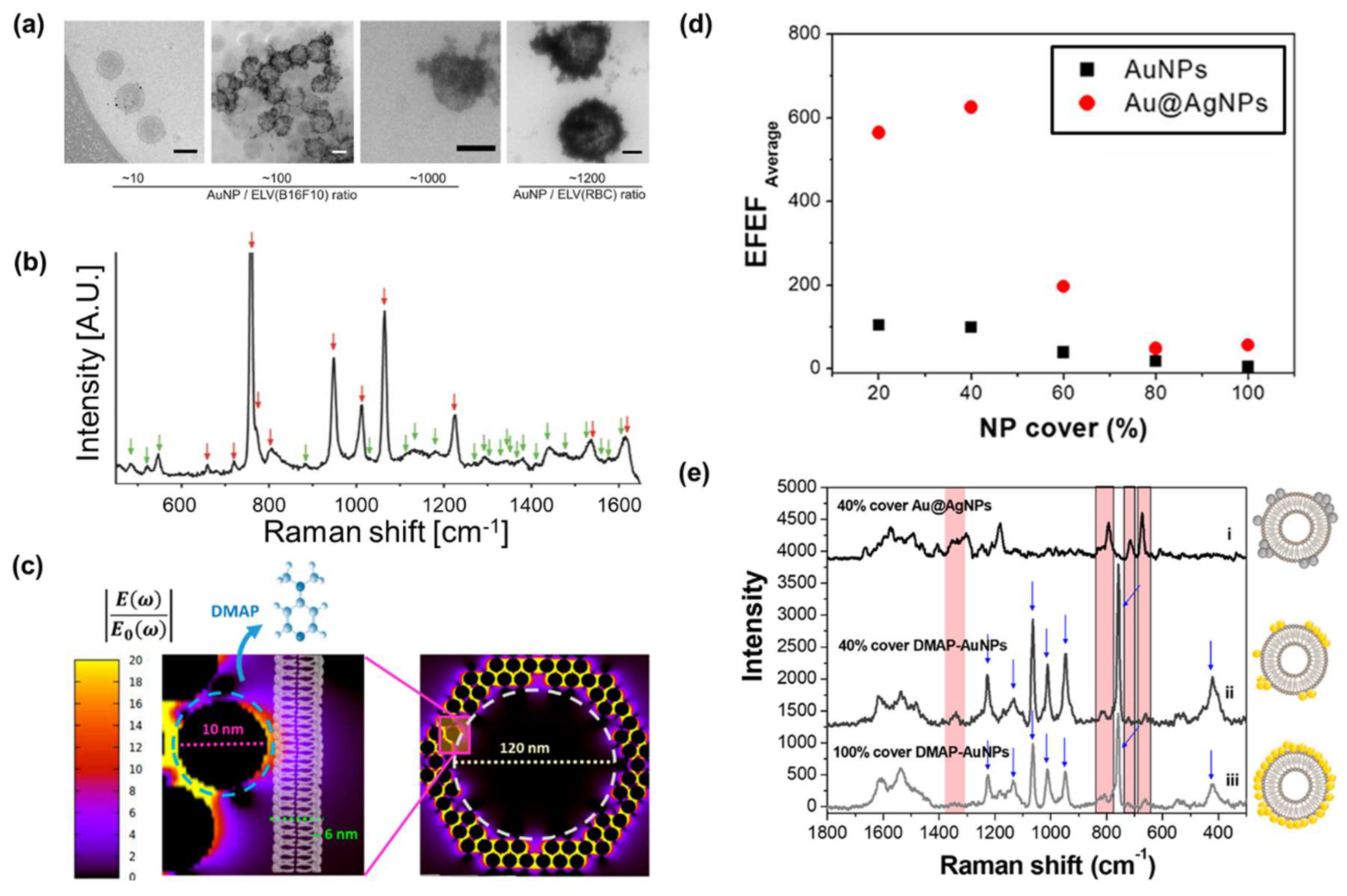
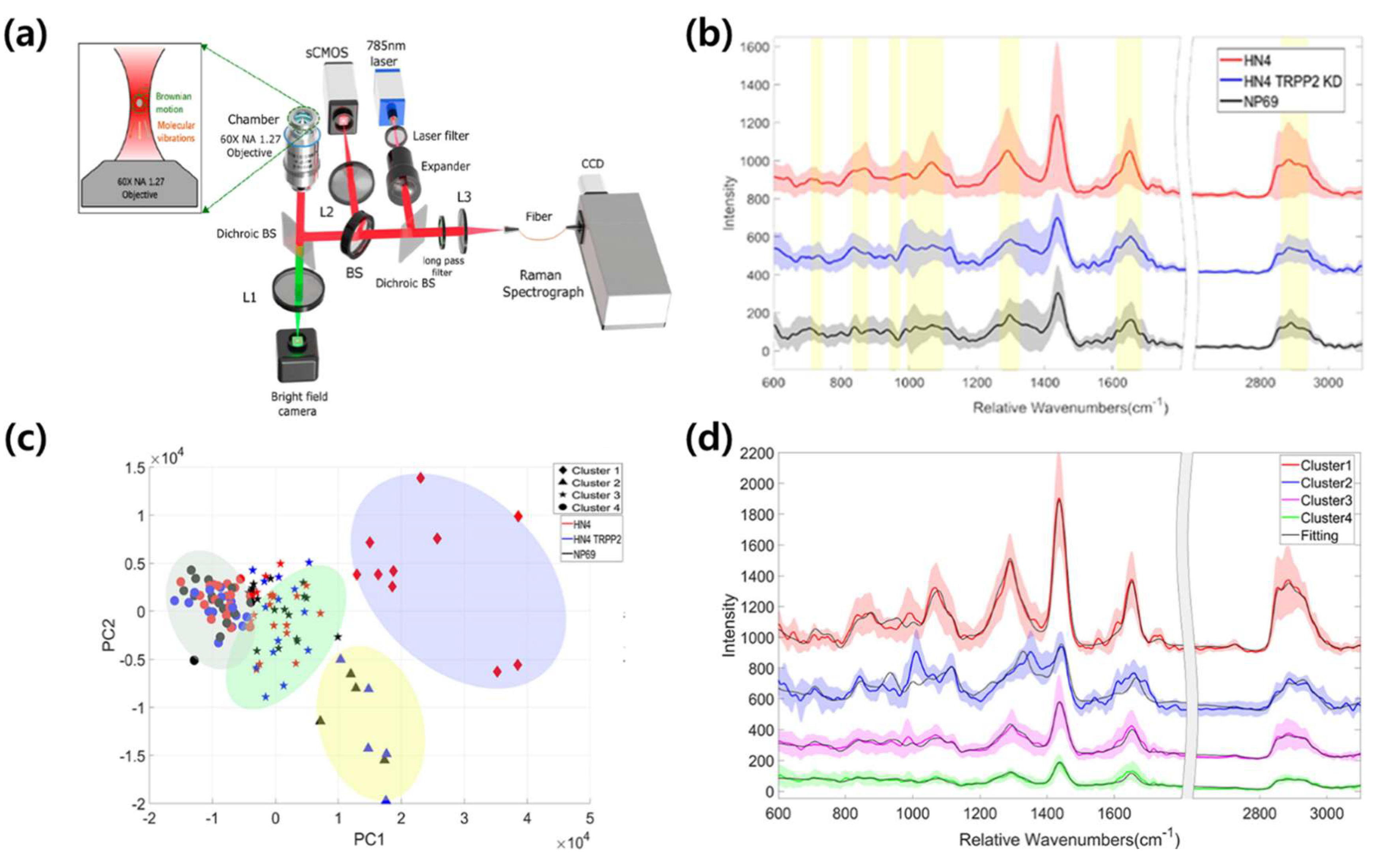
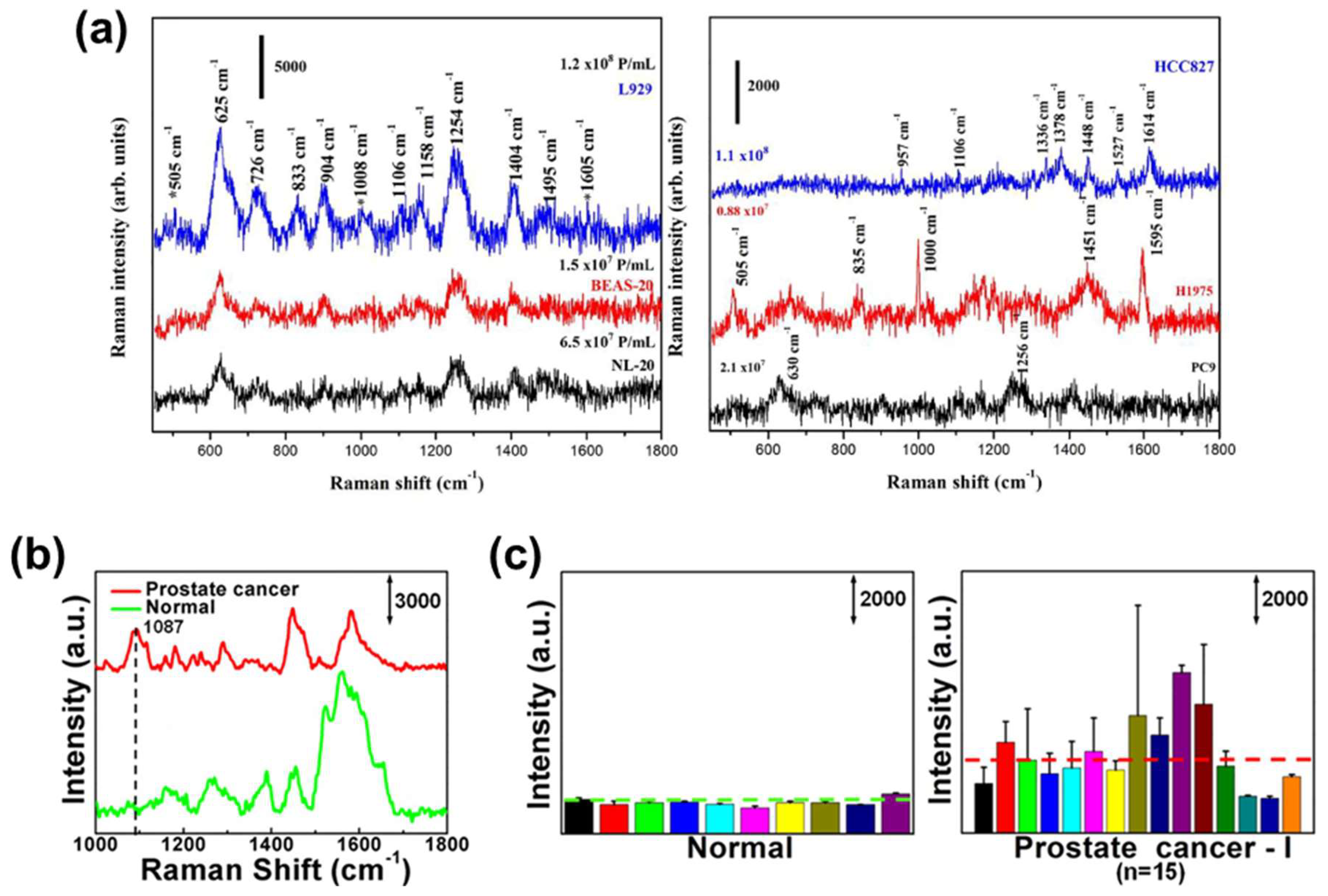

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, H.; Seo, D.; Choi, Y. Extracellular Vesicle Identification Using Label-Free Surface-Enhanced Raman Spectroscopy: Detection and Signal Analysis Strategies. Molecules 2020, 25, 5209. https://doi.org/10.3390/molecules25215209
Shin H, Seo D, Choi Y. Extracellular Vesicle Identification Using Label-Free Surface-Enhanced Raman Spectroscopy: Detection and Signal Analysis Strategies. Molecules. 2020; 25(21):5209. https://doi.org/10.3390/molecules25215209
Chicago/Turabian StyleShin, Hyunku, Dongkwon Seo, and Yeonho Choi. 2020. "Extracellular Vesicle Identification Using Label-Free Surface-Enhanced Raman Spectroscopy: Detection and Signal Analysis Strategies" Molecules 25, no. 21: 5209. https://doi.org/10.3390/molecules25215209




